通过shaplefile裁剪NetCDF
NetCDF(http://www.unidata.ucar.edu/software/netcdf/)格式是比较常见的地学数据存储格式,但是,很多NetCDF数据都是全球或大区域尺度,存储整个数据很浪费空间(比如中国区域CFMD数据集解压后占据硬盘380多G,而占国土面积1/4的青藏高原裁剪后的CFMD-QTP预计能节约90%的空间),网上找了很久也没有找到合适的裁剪code。本文描述了通过shapefile来裁剪Python的思路(需要netcdf4-python、gdal 等支持)。
1.效果
示例NetCDF下载地址http://westdc.westgis.ac.cn/data/7a35329c-c53f-4267-aa07-e0037d913a21
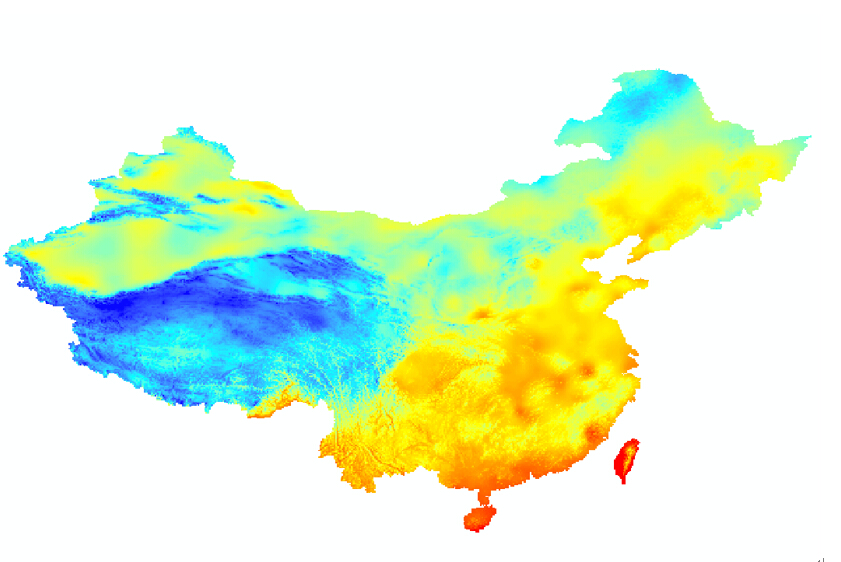
原始数据(130M):
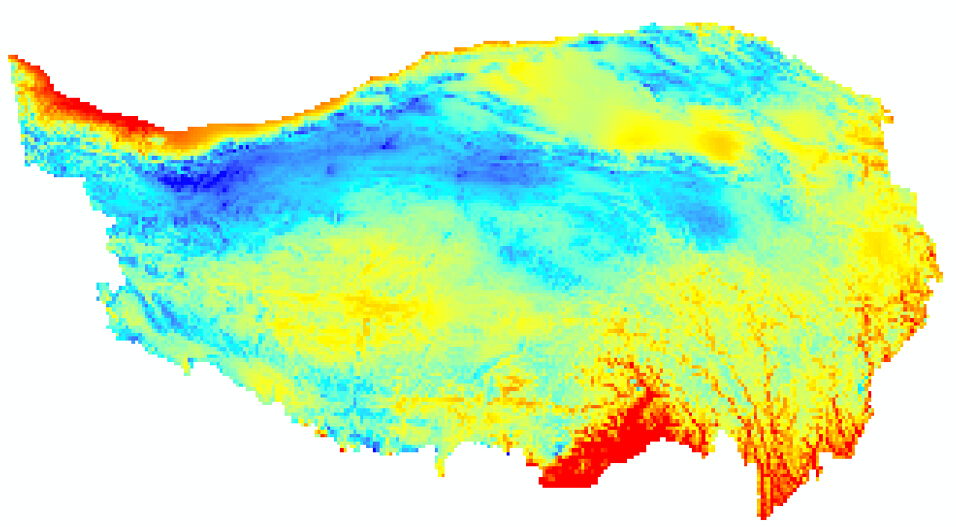
通过青藏高原边界clip后的数据(18M):
2.实现(需要PYTHON运行,数据量大时,建议python 64bit,支持批处理。)
具体思路如下:
- gdal计算shapefile 的 四角boundary 和所有点的boundary
- 其次根据四角boundary subset
- 在根据所有点boundary 计算netcdf mask并更新到QTP netcdf 的mask
- netcdf4-python将mask后netcdf的文件写出来 具体code如下:
##########################################################################################################
# NAME
# SubSetNetCDF.py
# PURPOSE
#
# PROGRAMMER(S)
# wuxb
# REVISION HISTORY
# 20150910 -- Initial version created and posted online
#
# REFERENCES
##########################################################################################################
import numpy as np
import numpy.ma as ma
import os
import formic
import datetime
from netCDF4 import Dataset
from osgeo import gdal, ogr
###################################################################################
# input data
###################################################################################
strShpFilePath = "D:\\Temp\\Shp\\Tibet_Plateau_Boundar.shp"
strNetCDFPathInput = "E:\\ForcingData\\DAT-31.12_NG_ITPCAS-CMFD_V0106_B-01\\Data_forcing_03hr_010deg_Unzip\\"
strNetCDFPathOutput = "D:\\workspace\\Data\\CFMD(QTP)\\"
###################################################################################
# Process
# read
# process
# write
###################################################################################
#read shapefile
shpDS = ogr.Open(strShpFilePath)
shpLyr = shpDS.GetLayer()
Envelop = shpLyr.GetExtent()
xmin,ymin,xmax,ymax = [Envelop[0],Envelop[2],Envelop[1],Envelop[3]] #Your extents as given above
mask_RES = []
#
fileset = formic.FileSet(include="**/*.nc", directory=strNetCDFPathInput)
nFile = 0 # print process file ID
########################################################################
#Function
#
########################################################################
def find_all_index(arr,item):
return [i for i,a in enumerate(arr) if a==item]
def getDimVar(ncInput,varList,varName):
varIdx = find_all_index(varList,varName)
if(len(varIdx) == 1):
varData = ncInput.variables[varName][:]
varList.remove(varName)
return varData
else:
print "Error there have no dim"
os.system("pause")
exit(0)
def getDataVar(ncInput,varList):
#check varlist
if(len(varList) == 1):
varName = str(varList[0])
varData = ncInput.variables[varName][:]
return varName,varData
else:
print "Error there have no dim"
os.system("pause")
exit(0)
########################################################################
#Process
########################################################################
for file_name in fileset:
nFile+=1
print "################################################################"
print "Current file is : " + file_name + "; It is the " + str(nFile)
print "################################################################"
#read netcdf lon lat data
ncInput = Dataset(file_name,'r', Format='NETCDF3_CLASSIC')
#list variables
varList = ncInput.variables.keys()
#get var
lon_Ori = getDimVar(ncInput,varList,'lon')
lat_Ori = getDimVar(ncInput,varList,'lat')
time = getDimVar(ncInput,varList,'time')
varDataName,varData_Ori = getDataVar(ncInput,varList)
########################################################################
#create mask
########################################################################
if len(mask_RES) == 0 :
#get boundary and xs ys
lat_bnds, lon_bnds = [ymin, ymax], [xmin, xmax]
lat_inds = np.where((lat_Ori > lat_bnds[0]) & (lat_Ori < lat_bnds[1]))
lon_inds = np.where((lon_Ori > lon_bnds[0]) & (lon_Ori < lon_bnds[1]))
ncols = len(lon_inds[0])
nrows = len(lat_inds[0])
#create geotransform
xres = (xmax - xmin) / float(ncols)
yres = (ymax - ymin) / float(nrows)
geotransform = (xmin,xres,0,ymax,0, -yres)
#create mask
mask_DS = gdal.GetDriverByName('MEM').Create('', ncols, nrows, 1 ,gdal.GDT_Int32)
mask_RB = mask_DS.GetRasterBand(1)
mask_RB.Fill(0) #initialise raster with zeros
mask_RB.SetNoDataValue(-32767)
mask_DS.SetGeoTransform(geotransform)
maskvalue = 1
err = gdal.RasterizeLayer(mask_DS, [maskvalue], shpLyr)
mask_DS.FlushCache()
mask_array = mask_DS.GetRasterBand(1).ReadAsArray()
mask_RES = ma.masked_equal(mask_array, 255)
ma.set_fill_value(mask_RES, -32767)
########################################################################
#subset
########################################################################
var_subset = varData_Ori[:,min(lat_inds[0]):max(lat_inds[0]) + 1, min(lon_inds[0]):max(lon_inds[0]) + 1]
var_subset._set_mask(np.logical_not(np.flipud(mask_RES.mask))) # update mask (flipud is reverse 180)
lon_subset = lon_Ori[lon_inds]
lat_subset = lat_Ori[lat_inds]
###################################################################################
# Open a new NetCDF file to write the data to. For format, you can choose
# from
# 'NETCDF3_CLASSIC', 'NETCDF3_64BIT', 'NETCDF4_CLASSIC', and 'NETCDF4'
###################################################################################
#create file path and name
InputFileDir,InputFile = os.path.split(file_name)
InputDir,InputFileDirName = os.path.split(InputFileDir)
OutputFileDir = os.path.join(strNetCDFPathOutput,InputFileDirName)
if not os.path.exists(OutputFileDir):
os.makedirs(OutputFileDir)
strInputFileList = InputFile.split('_')
strInputFileList[1] = strInputFileList[1] + "-QTP"
OutfileName = '_'.join(strInputFileList)
OutputFileDirName = os.path.join(OutputFileDir,OutfileName)
#create file
ncOutput = Dataset(OutputFileDirName, 'w', format='NETCDF3_CLASSIC')
ncOutput.description = "Extract Data %s from CFMD by QTP shapefile. %s" % \
(ncInput.variables[varDataName].long_name.lower(),ncInput.description)
# Using our previous dimension info, we can create the new time dimension
# Even though we know the size, we are going to set the size to unknown
ncOutput.createDimension('time', None)
ncOutput.createDimension('lon', ncols)
ncOutput.createDimension('lat', nrows)
# Add lon Variable
var_out_lon = ncOutput.createVariable('lon', ncInput.variables['lon'].dtype,('lon',))
for ncattr in ncInput.variables['lon'].ncattrs():
var_out_lon.setncattr(ncattr, ncInput.variables['lon'].getncattr(ncattr))
ncOutput.variables['lon'][:] = lon_subset
# Add lat Variable
var_out_lat = ncOutput.createVariable('lat', ncInput.variables['lat'].dtype,('lat',))
for ncattr in ncInput.variables['lat'].ncattrs():
var_out_lat.setncattr(ncattr, ncInput.variables['lat'].getncattr(ncattr))
ncOutput.variables['lat'][:] = lat_subset
# Add time Variable
var_out_time = ncOutput.createVariable('time', ncInput.variables['time'].dtype,('time',))
for ncattr in ncInput.variables['time'].ncattrs():
var_out_time.setncattr(ncattr, ncInput.variables['time'].getncattr(ncattr))
ncOutput.variables['time'][:] = time
# Add data Variable
var_out_data = ncOutput.createVariable(varDataName, ncInput.variables[varDataName].dtype, ("time","lat","lon",))
for ncattr in ncInput.variables[varDataName].ncattrs():
var_out_data.setncattr(ncattr, ncInput.variables[varDataName].getncattr(ncattr))
ncOutput.variables[varDataName][:] = var_subset
# attr
ncOutput.history = "CLIP Created datatime" + datetime.datetime.now().strftime("%Y-%m-%d %H:%M:%S") + " by CAREERI wuxb"
ncOutput.source = "netCDF4 1.1.9 python"
###################################################################################
# write close
###################################################################################
# close
ncOutput.close() # close the new file
ncInput.close() # close ori file
print 'Done'
print 'End time: ' + datetime.datetime.now().strftime("%Y-%m-%d %H:%M:%S")